Reviewed by Paul Domanski
In order for the cell to function normally, a balance must be maintained between the production, degradation, and clearance of cytoplasmic components. To accomplish this, the cell relies primarily on two methods: 1) the ubiquitin-proteasome pathway, in which proteins are selectively tagged by ubiquitin for degradation in the proteasome and 2) autophagy (‘selfeating’), a general term used to describe all pathways that are used to deliver cytoplasmic components to the lysosome for degradation. While the ubiquitin-proteasome pathway is mainly used to degrade short-lived and abnormal proteins, autophagy is responsible for elimination of cytoplasmic components, damaged organelles, and long-lived and aggregated proteins. This cellular ‘recycling’ system is tightly regulated so that degradation and regeneration of the cellular building blocks can proceed in an efficient manner.
Ubiquitin
Ubiquitination, one of the most common post-translational modifications (PTMs) in the cell, is the primary mechanism by which short-lived proteins are targeted for degradation and clearance. It is a highly specific system in which ubiquitin, a small (76 amino acid) protein, forms an amide bond with an epsilon amine of lysine in the target protein. This happens in a three-step enzymatic process:
- Activation of ubiquitin by E1 (ATP-dependent).
- Transfer of ubiquitin from E1 to the active site cysteine of a ubiquitin-conjugating (UBC) enzyme E2 (more than 30 UBC proteins are known).
- Ligation of the C-terminal glycine of ubiquitin to the target protein by E3. The E3 family of ubiquitin ligases currently numbers more than 500 members.

Figure 1. The Ubiquitin-Proteasome Pathway Target proteins are ubiquitinated in a three-step ATP-dependent process involving E1, E2, and E3. The protein is then unfolded and degraded into small peptides in the proteasome, where they can be used in antigen presentation or hydrolyzed to individual amino acids.
In the ubiquitination cascade, E1 can bind with dozens of E2s, which can bind with hundreds of E3s in a hierarchical way. Furthermore, proteins can be monoubiquitinated, multiubiquitinated, and polyubiquitinated, which adds a certain depth to the pathway, as different types of linkages can activate different signaling pathways, through binding to specific ubiquitin binding domains (more than 20 have been discovered). Similar to phosphorylation, ubiquitination is reversible, and ∼85 deubiquitinases (DUBs) are known. The majority of ubiquinated proteins are destined for degradation by the proteasome, however, these modifications can also target proteins to particular destinations in the cell as well as mediate protein-protein interactions (Figure 1).1-3
Ubiquitination is involved in numerous cell functions and pathologies, including immune response, DNA repair, signal
transduction, cancer, neurological disorders, and more. Obviously, any defects in the pathway can have widespread
effects in the cell and are implicated in a range of disease states including neurodegeneration, cancer, and cardiovascular disease.4,5 With the advent of new reagents and technology (e.g., mass spectrometry), greater focus is placed on developing drugs that target the ubiquitin-proteasome pathway.
Cayman has an extensive product portfolio geared toward the study of ubiquitin and the proteasome. Numerous chemicals that can enhance or block the effects of ubiquitination are available to allow specific pathways in the ubiquitination process to be analyzed. We also have a panel of antibodies, including FK1 and FK2, which can be used to screen for particular types of ubiquitination of target proteins (monoubiquitination, multiubiquitination, and polyubiquitination). We have recently added kits that can be used to test for ubiquitination, ubiquitin binding proteins, and deubiquitinase activity, giving researchers key tools needed to further investigate the processes involved and to discover compounds that influence these pathways.
Autophagy
Autophagy is an evolutionarily conserved catabolic process in which cellular components are degraded through the lysosomal machinery. As such, it is a normal part of cell growth, development, differentiation, and homeostasis. This enables the cell to maintain a strict balance between synthesis, degradation, and recycling of cellular components. In lesser organisms, its purpose is to maintain the metabolic equilibrium with ever-changing nutrient availability.6 Although originally thought to be a ‘crude’ process for the destruction of cell constituents, it has become apparent that there is a ‘selective’ aspect to autophagy. In higher organisms, this system can eliminate aggregated proteins, damaged organelles such as mitochondria (mitophagy) and peroxisomes (pexophagy), and can also remove microbial pathogens (xenophagy) (Figure 2).7,8 Autophagy is continuously operating at a basal level in cells, but is also highly inducible in response to internal and external stimuli. Numerous signaling pathways are involved in autophagy, though to what extent is not well understood. Many proteins have been implicated in the process of autophagy, including mTOR, p62, Beclin 1, and PINK1 (Figure 3).8 Due to the balancing act that autophagy must perform, one would expect that any defects in the system could play an important role in disease. Indeed, it has been shown that knockout mice lacking an essential gene for autophagy will die within hours of birth.9 It is also known that disruptions in autophagy have a role in cancer, neurodegenerative diseases (both Huntington’s and Alzheimer’s disease cells show increased autophagic vacuole accumulation), lysosomal storage diseases, infectious diseases and DNA damage response (through HDACs) and may play a role in aging and cell death (through mitophagy).10,11

Figure 2. Nonselective and Selective Autophagy (A) Under starvation conditions, nonselective autophagy catabolizes components in the cytoplasm into basic building blocks that the cell machinery can reuse. (B) Certain stimuli can drive selective autophagy, as in stimulation of p62, damaged mitochondria, or infection.
The hallmark of autophagy is the presence of a unique organelle called the autophagosome, a double membranebound vacuole that can range in size from 300 to 900 nm. Autophagosomes arise from a single membrane structure known as the phagophore, which elongates and then closes in on itself, sequestering cytoplasmic material in the process. These fully enclosed phagosomes then fuse with the lysosome to form the autolysosome, which allows for degradation of the components.6
In the late 1990s, studies in yeast led to the elucidation of a set of more than 20 autophagy-related genes (ATGs), and further research led to the discovery of mammalian homologs for many of these genes and their associated proteins.12,13 A schematic representation of the signaling cascade, as well as known inhibitors and activators, is shown in Figure 3. Under nutrientrich conditions, the protein kinase mTOR (mammalian target of rapamycin) is a primary negative regulator of autophagy. However, in a starvation situation, mTOR is deactivated, which allows ULK (the mammalian counterpart of the yeast protein ATG1) to become activated and translocate to the endoplasmic reticulum, initiating the autophagy cascade. The formation of the phagosome concludes when ATG8 (in mammals, microtubuleassociated protein light chain 3, (MAP-LC3 or LC3-I)) is cleaved in a ubiquitin-like process at the C-terminus by ATG4 followed by the addition of a phosphatidylethanolamine (PE) moiety to ATG8. This LC3-phospholipid (LC3-II) covalently associates with the phagophore and is essential for the formation of the phagosome. As such, LC3-II serves as an excellent marker for detection of the autophagosome. It is worth noting that this process, as with ubiquitination, is reversible, with ATG4 also playing a role in the delipidation reaction.

Figure 3. Autophagy Pathway in Mammals (A) Prototypical induction of autophagy by mTOR (inhibited by rapamycin) that leads to the (B) Nucleation/expansion of the autophagosomal membrane using a protein complex known to contain Beclin 1 and PI3K (inhibited by 3-methyladenine (3-MA) and wortmannin). (C) Expansion of the membrane occurs via a ubiquitin like mechanism, wherein ATG7 and ATG3 convert LC3-I to LC3-II, which localizes to the autophagosome membrane.
Several methods exist for analyzing the extent of autophagy in cells. Early attempts relied on measuring increases in the numbers of autophagosomes by staining with acidotropic dyes such as monodansylcadaverine (MDC) (Cayman’s Autophagy/ Cytotoxicity Dual Staining Kit) and transmission electron microscopy (TEM), the first detection method for autophagy.14
Interestingly, the phospholipid modification directed to LC3 causes an elecrophoretic mobility shift, which can be used to determine the levels of LC3-I and LC3-II by analysis on western blot. When this technique is applied in concert with known lysosomal inhibitors, such as Bafilomycin A1 , the accumulation of LC3-II in the autophagosome-lysosome can be measured, which allows one to ascertain ‘autophagic flux’, the rate at which proteins and cellular material are cleared through the autophagic process.
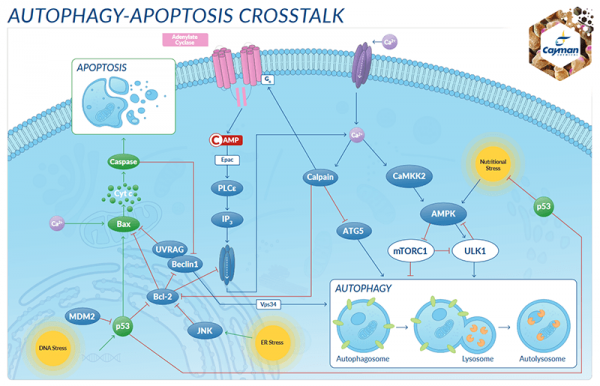
The ubiquitin-proteasome and autophagy pathways are vital to the cell’s ability to maintain the delicate balance between the production and degradation of proteins and cellular components. It is now understood that these pathways can also involve ‘cross-talk’, and that autophagy can act as an additional method for eliminating ubiquitinated proteins under conditions of extreme stress. Although much is known, much more research must be accomplished to elucidate improved biomarkers for determining how these pathways can be utilized in relevant disease settings to address and monitor therapeutic drug effectiveness.
References
- Husnjak, K. and Dikic, I. Annu. Rev. Biochem. 81, 291-322 (2012).
- Komander, D. and Rape, M. Annu. Rev. Biochem. 81, 203-209 (2012).
- Spasser, L. and Brik, A. Agnew. Chem. Int. Ed. Engl. 51(28), 6840-6862 (2012).
- Fulda, S., Rajalingam, K., and Dikic, I. EMBO Mol. Med. 4(7), 545-556 (2012).
- Shaid, S., Brandts, C.H., Serve, H., et al. Cell Death Differ. 20(1), 21-30 (2013).
- Abeliovich, H. and Klionsky, D.J. Microbiol. Mol. Biol. Rev. 65(3), 463-479 (2001).
- Jin, M., Liu, X., and Klionsky, D.J. Cell 152(1-2), 368 (2013).
- Mizushima, N. and Komatsu, M. Cell 147(4), 728-741 (2011).
- Komatsu, M., Waguri, S., Ueno, T., et al. J. Cell Biol. 169(3), 425-434 (2005).
- Todde, V., Veenhuis, M., and van der Klei, I.J. Biochim. Biophys. Acta 1792(1), 3-13 (2009).
- Robert, T., Vanoli, F., Chiolo, I., et al. Nature 471(7336), 74-79 (2011).
- Klionsky, D.J., Cregg, J.M., Dunn, W.A., Jr., et al. Dev. Cell 5(4), 539-545 (2003).
- Legakis, J.E. and Klionsky, D.J. Deretic, V., editor, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 1-17 (2006).
- Klionsky, D.J., Abdalla, F.C., Abeliovich, H., et al. Autophagy 8(4), 445-544 (2012).
- Mizushima, N. and Yoshimori, T. Autophagy 3(6), 542-545 (2007).
- Mizushima, N., Yoshimori, T., and Levine, B. Cell 140(3), 313-326 (2010).
- Rubinsztein, D.C., Mariño, G., and Kroemer, G. Cell 146(5), 682-695 (2011).