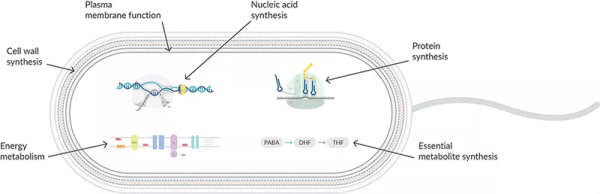
Protein synthesis inhibitors act at the ribosome level, working at different stages of prokaryotic mRNA translation into proteins (Figure 1). They are highly selective to 70S ribosomes in prokaryotic cells since eukaryotic cells have a different ribosomal size, sequence, structure, and ratio of protein to RNA. Translation in prokaryotes involves the assembly of a small subunit (30S) and a large subunit (50S) to form a ribosome that binds mRNA to read the nucleotide sequence. tRNA binds to the A, P, and E sites during the initiation, elongation, and termination stages to translate the polypeptide sequence. By targeting different stages of mRNA translation, antibiotics can be swapped if resistance develops.

Figure 1: Antibiotics interfere with multiple stages of protein synthesis.
Tetracyclines bind reversibly to the 30S ribosomal subunit at a position that blocks the binding of the tRNA to the acceptor site on the mRNA-ribosome complex. Resistance to tetracyclines develops primarily by preventing accumulation of the drug inside the bacterial cell either by decreasing influx or increasing efflux.
Aminoglycosides and aminonucleosides, such as puromycin, bind to the active site of 16S rRNA within the 30S ribosomal subunit, interfering with the accurate recognition of tRNA by rRNA during translation. The initial steps required for peptide synthesis, such as binding of mRNA and the association of the 50S ribosomal subunit, are uninterrupted. The elongation fails to occur due to disruption of the mechanisms for ensuring translational accuracy, causing premature termination of nascent polypeptide chains. In the case of puromycin, these properties have been exploited in assays for peptide-bond formation and elongation and also as a selection agent in cell culture systems. Resistance to aminoglycosides develops from structural modifications to the antibiotic made by aminoglycoside phosphotransferases, aminoglycoside nucleotidyltransferases, and aminoglycoside acetyltransferases expressed in resistant organisms.
Oxazolidinones bind at the P site of the ribosomal 50S subunit, inhibiting the binding of tRNA and preventing the formation of the initiation complex. If the 70S complex is already formed, then binding of oxazolidinones inhibits translocation of the peptide chain from the A site to the P site during formation of the peptide bond. Mutations at the 23S rRNA within the 50S ribosomal subunit can lead to a diminished affinity for oxazolidinones, which may cause resistance.
Amphenicols block the peptidyl transfer step of elongation on the 50S subunit in bacteria, most likely acting at the A site. They can also perturb additional ribosomal functions, such as termination, translational accuracy, and synthesis of 50S ribosomal subunits. Chloramphenicol can also bind the mitochondrial 70S ribosome, inhibiting mitochondrial protein synthesis and thus producing toxicity in eukaryotes. Mutations or alterations to the target of amphenicols are a common mechanism of resistance.
Lincosamides act as structural analogs of portions of tRNA during the initial phase of pretranslocation in the peptide elongation phase, interrupting the process of peptide chain initiation in the 50S subunit. They can also cause peptidyl-tRNA to dissociate from ribosomes. Resistance to lincosamides may be caused by methylation of the 23S rRNA, modification of the antibiotics by specific enzymes, or active efflux systems.
Most macrolides interact with 23S rRNA within the 50S ribosomal subunit, blocking the approach to the elongating peptide's exit tunnel, which results in a premature release of peptidyl-tRNA intermediates. They compete with lincosamide antibiotics that are direct peptidyl transferase inhibitors. The three main mechanisms responsible for acquired macrolide resistance include ribosomal target modification, an active drug efflux mechanism, and mutation in the 23S rRNA.
Streptogramins act in a similar way to lincosamides and macrolides depending on type. Type A streptogramins block substrate attachment to the donor and acceptor sites of the peptidyl transferase center on the 50S subunit, preventing the earliest event of elongation. Type B streptogramins compete with macrolides for binding the 50S ribosome to interfere with the formation of long polypeptides and cause a premature detachment of incomplete peptide chains. Type A streptogramins can increase the ribosome affinity for type B streptogramins, causing a bactericidal synergy between the two normally bacteriostatic agents. Resistance to these antibiotics mainly involves inactivation of inhibitors and mutations of ribosomal proteins or rRNA genes.
Nitrofurans have a unique mechanism for slowing bacterial growth. They are prodrugs that require reductive activation by two redundant bacterial flavoproteins to reactive intermediates. These reactive intermediates have diverse effects: triggering DNA lesions, inducing oxidative stress, and inhibiting RNA and protein synthesis. It is not known if the reactive intermediates directly modify DNA, RNA, and proteins or if they primarily target the cellular machineries that carry out replication, transcription, translation, and energy metabolism. Though rare, mutations in genes encoding the prodrug-activating enzymes can lead to resistance.