β-Lactam antibiotics, including penicillins, cephalosporins, monobactams, and carbapenems, are distinguished by a lactam ring in their molecular structure and act by inhibiting the synthesis of the peptidoglycan layer of bacterial cell walls. They interfere with the transpeptidase, transglycosylase, and/or carboxypeptidase activities of species-specific membrane-bound penicillin-binding proteins (PBPs) that facilitate cross-linking of the cell wall components during the final stage of bacterial cell wall synthesis (Figure 1). This occurs by binding to the terminal D-Ala-D-Ala in the lengthening peptidoglycan structure and rapidly acylating the active site serine of the PBP, followed by very slow deacylation, which inactivates the PBP. As the cell grows, it is not able to synthesize more cell wall to accommodate the expansion. As a result, the pressure inside the cell will push the plasma membrane out of a weak spot in the cell wall like a balloon, which will eventually rupture. Because the formation of a division furrow to create new daughter cells depends on the ability to synthesize a new cell wall, the cell is unable to pinch off the extra cytoplasmic material. An extremely vulnerable spheroplast is formed when the cell wall sheds entirely. In this form, bacteria lose control over their shape, and the duplication of much of their genetic and metabolic material further disrupts homeostasis, which leads to cell death.
Bacteria develop resistance to β-lactam antibiotics by synthesizing β-lactamase, an enzyme that attacks the β-lactam ring to inactivate the antibiotic. More than 1,000 unique β-lactamases have been described. To overcome this resistance, β-lactam antibiotics are usually given with β-lactamase inhibitors. Efflux and modification or deletion of porin function can also play a role in resistance to some β-lactams in conjunction with β-lactamase production, which results in resistance to virtually all β-lactams. Some bacteria area also able to acquire new PBPs that have low affinity for β-lactams. The best-known example of this strategy for resistance occurs with methicillin-resistant S. aureus (MRSA) that evolved a PBP with low affinity for all but a few recently developed β-lactams.

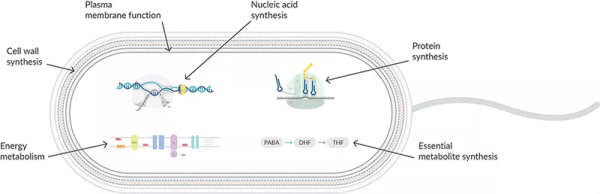
Figure 1: Antibiotics interfere with various aspects of the synthesis of the peptidoglycan cell wall. Gram-negative cell walls consist of two layers external to the cell membrane—a thin layer of peptidoglycan and an outer membrane with porins that allow certain antibiotics but not glycopeptides to diffuse through to their site of action. Gram-positive bacteria have a thicker peptidoglycan layer but lack an extra outer membrane enabling glycopeptide access.
Glycopeptides and lipoglycopeptides are composed of glycosylated cyclic or polycyclic nonribosomal peptides that interfere with cell wall formation by forming a complex between the antibiotic and the C-terminal D-Ala-D-Ala dipeptide of the nascent peptidoglycan on the outer surface of the cell membrane of Gram-positive bacteria (Figure 1). The formation of this complex prevents the transglycosylation and transpeptidation reactions that are necessary for completion of the peptidoglycan chain, resulting in an incomplete cell wall and subsequent cell death. They are too bulky to pass through the porin channels found in the outer membrane of Gram-negative bacteria. In addition to its interaction with D-Ala-D-Ala, some antibiotics in this group bind to lipid II, a cell wall precursor on the cytoplasmic side of the cell membrane that must translocate across the cell membrane to deliver and incorporate its disaccharide-pentapeptide monomer for cross-linking into peptidoglycan. This interaction results in membrane depolarization and eventual membrane disruption that leads to cell death. Since glycopeptides target the outer surface of the cell wall, they do not have to overcome a membrane barrier and do not interact with enzymes. Known glycopeptide resistance mechanisms instead are associated with structural modifications in the substrates for the enzymes that incorporate the final amino acids in the pentapeptide precursors. The most relevant modifications involve replacement of the C-terminal D-Ala with D-lactate or D-serine, resulting in D-Ala-D-Lac or D-Ala-D-Ser peptide sequences with reduced binding affinities for the antibiotic.
The phosphonic acid fosfomycin inhibits the growth of a wide variety of bacteria by targeting the enolpyruvyl transferase MurA, which catalyzes the first committed step of peptidoglycan synthesis that is essential for cell wall formation (Figure 1). It enters cells through glycerol-3-phosphate and hexose-6-phosphate transporters. Deficiency in these transporters that cause increased efflux or decreased cellular uptake can lead to resistance. Specific murA gene mutations that produce an enzyme with reduced affinity for fosfomycin or increased expression of MurA that overwhelms the capacity of fosfomycin for growth inhibition are also known causes of resistance. Fosfomycin-inactivating enzymes have also been documented.